Interamerican Journal of Heath Sciences 4 (2024) - ISSN 2953-3724
In silico exploration of potential breast cancer drugs
Exploración in silico de fármacos potenciales contra el cáncer de mama
Daneilys de Dios Hernández1 *
1Universidad de Ciencias Médicas Pinar del Río.
![]()
Received: 28-06-2023 Revised: 05-09-2023 Accepted: 03-01-2024 Published: 04-01-2024
How to Cite: de Dios Hernández D. In silico exploration of potential breast cancer drugs. Interamerican Journal of Health Sciences. 2024; 4:171. https://doi.org/10.59471/ijhsc2024171
Introduction: breast carcinoma is the most common neoplasm among women. One of the current problems is the emergence of drug resistance. Cathepsin B is a cysteine protease that is overexpressed in tumor tissue. The search for new therapeutic alternatives derived from the nitroxoline nucleus constitutes an encouraging solution against the disease.
Objective: to evaluate in silico potential cathepsin B inhibitors as therapeutic targets in the treatment of breast carcinoma.
Methods: from the PubChem database, 12 ligands derived from the nitrosoline nucleus were obtained, which were converted to 3D and their energy was minimized by applying the GAFF force field. The specific characteristics of the protease active site were determined with the Proteins Plus web server to carry out molecular docking studies with Autodock Tools.
Results: ligands 1511784, 14125599 and 45487202 showed a favorable affinity energy (ΔG=-5,56; -5,51 and -5,08 respectively) and inhibition constant by inhibiting key residues of the catalytic site of cathepsin B. The amino acids were: His199, His110, His111, Gln23 and Gly198. The main interaction was by hydrogen bonding.
Conclusions: nitrosoline contains anticancer properties. Ligands 1511784, 14125599 and 45487202 constitute potential drugs against breast cancer. Therefore, in silico analyzes reduce the cost of current research and contribute to the specificity and immunogenicity of therapies and pharmacological biosafety.
KEYWORDS
Nitrosoline, Cathepsin B Inhibitors, Molecular Docking.
RESUMEN
Introducción: el carcinoma de mama es la neoplasia más frecuente entre las mujeres. Uno de los problemas actuales es la resistencias a fármacos. La catepsina B es una cisteíno proteasa que se sobre expresa en el tejido tumoral. La búsqueda de nuevas alternativas terapéuticas derivadas del núcleo de la nitrosolina constituye una solución contra la enfermedad.
Objetivo: evaluar in silico inhibidores potenciales de la catepsina B como blancos terapéuticos en el tratamiento del carcinoma mamario.
Métodos: a partir de la base de datos PubChem se obtuvieron 12 ligandos derivados del núcleo de la nitrosolina que fueron convertidos a 3D y se les minimizó la energía a partir de la aplicación del campo de fuerza GAFF. Se determinaron las características del sitio activo de la proteasa con el servidor web Proteins Plus para la realización de los estudios de acoplamiento molecular con Autodock Tools.
Resultados: los ligandos 1511784, 14125599 y 45487202 mostraron una energía de afinidad (ΔG=-5,56; -5,51 y -5,08 respectivamente) y constante de inhibición favorable al inhibir residuos claves del sitio catalítico de la catepsina B. Los aminoácidos fueron: His199, His110, His111, Gln23 y Gly198. La principal interacción fue por enlace de hidrógeno.
Conclusiones: la nitrosolina contiene propiedades anticancerígenas. Los ligandos 1511784, 14125599 y 45487202 constituyen fármacos potenciales contra el cáncer de mama. Por ello, los análisis in silico reducen el costo de las investigaciones y contribuyen a la especifidad e inmunogenicidad de las terapias y la bioseguridad farmacológica.
PALABRAS CLAVES
Nitrosolina, Inhibidores de la Catepsina B, Acoplamiento Molecular.
INTRODUCTION
Breast carcinoma (BC) is the most frequent neoplasm among women. It is the second leading cause of cancer death among women, after lung cancer.(1) It is estimated that 30 % of breast neoplasms are due to modifiable risk factors, such as excess body weight, physical inactivity, and alcohol intake.(2) It occurs when the cells of the mammary gland grow in an uncontrolled manner due to the failure of the regulatory mechanisms that control their proliferation.(3) Breast cancer incidence rates between 2010 and 2019 increased by 0.5 % annually.(2) According to the statistical yearbook, in 2019, 4351 new cases of MC were registered in Cuba. The age group most affected by breast carcinoma was between 60-79 years old, with 804 deaths, which confirmed that the older the age, the higher the incidence rate of this condition.(4)
The genetic variants associated with an increased risk of MC involve mutations in the BRCA1/2 genes. The most widely used therapy includes the selective estrogen receptor modulators tamoxifen and raloxifene, or the aromatase inhibitors, anastrozole, letrozole, and exemestane, to treat postmenopausal women aged 35 years or older.(1) Up to 80 % of invasive breast cancers are infiltrating ductal carcinomas (IDC).(5)
One of the current problems in oncology is the emergence of resistance to treatment, where malignant cells become insensitive to drugs.(6) Currently, the use of computational tools applied to drug design (in silico) allows the search for new therapies not explored in experimental design. This makes possible the selectivity and efficacy of a therapy, which reduces the time and cost of present research.
Cathepsin B is a lysosomal cysteine-type protease and plays an important role in intracellular proteolysis. Its overexpression has been observed in esophageal, gastric, prostate, glioblastoma, and breast cancer. Therefore, it is considered a potential target with therapeutic properties due to its wide participation in cancer progression.(7,8) Currently, work is being done on the design of cathepsin B inhibitors, and a reduction in tumorigenesis has been observed. The use of nitrosoline-derived inhibitors constitutes potential drugs in the treatment of tumors.(9) The following compounds derived from the nitroso line nucleus are being worked on, as well as molecular docking studies that allow the selection of possible candidates for the treatment of mammary carcinoma.
METHODS
A search for nitroxoline-derived and nitroxoline-like compounds was performed in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). A total of 19 compounds, 93 substances, and 390 structurally related compounds were found. Subsequently, filtering was performed in PubChem, taking into account characteristics such as molecular weight, number of carbon atoms, number of rotations, and the year when the compound was annotated. For this last parameter, the last 5 years were chosen. As a result, 30 compounds with the desired characteristics were obtained.
The data obtained in 2D was converted to 3D (three-dimensional structure of the compound) using the Avogadro program, version 1.2.0. The geometry of each compound was optimized, and the General AMBER Force Field (GAFF) was applied. This type of force field is compatible with organic molecules with C, H, O, N, S, P, F, Cl, and Br in their structure. Of the 30 compounds analyzed, those with an energy stability value greater than 100 KJ/mol were discarded. As a result, the data of substances was reduced to 12 compounds. Molecular visualization of each compound was possible with PoV Ray 3.7 software.
The 3D structure of the receptor protein of interest was taken from the Protein Data Bank (PDB, https://www.rcsb.org/). The 8b5f structure was chosen. The Proteins Plus web server (https://proteins.plus), a computational tool for the study of ligand-protein interactions, was used. This allowed the selection of the size of the cathepsin B binding pocket, the potential amino acid residues of the active site, and their possible interactions for molecular docking studies.
Next, the protein was separated from its ligands and water molecules for processing using the Pymol 1.74 program. Subsequently, molecular docking studies were performed to estimate the energetic interaction between cathepsin B and each of the ligands using Autodock Tools 1.5.7.
RESULTS
The compounds derived from the nitrosoline nucleus extracted from the PubChem database and converted to 3D with the Avogadro software are shown below (Figure 1). For each compound, the strength between its atoms was estimated, which is evidenced by a potential energy value assigned to each. The energy is given in KJ/mol (Table 1). Additionally, the table contains the chemical formula and molecular weight of each substance, as well as the number of atoms and bonds present in each one.

Fig 1. Nitrosoline-derived compounds extracted from the PubChem database. The PoV Ray program was used for 3D molecular visualization. Twelve compounds are shown in bar and sphere representation. Carbon atoms (C) are shown in gray, hydrogens (H) in white, oxygen (O) in red, nitrogen (N) in dark blue, chlorine (Cl) in green, and fluorine (F) in light blue.
|
Table 1. Compounds extracted from PubChem. Energy values calculated with the Avogadro 1.2.0 program are included. The GAFF force field was applied |
||||||
|
Linking |
Name |
Molecular formula |
Energy (KJ/mol) (GAFF) |
Molecular weight (g/mol) |
No. atoms |
No. links |
|
250068325 |
Nitrosoline |
C11H8N2O4 |
64,98 |
232,19 |
25 |
26 |
|
199506 |
5-phenyl azo-8-hydroxyquinoline |
C15H11N3O |
62,75 |
249,267 |
30 |
32 |
|
245295 |
5,7-Dinitro-8-quinolinolol |
C9H5N3O5 |
14,92 |
235,153 |
22 |
23 |
|
416002 |
5-Amino-8-hydroxyquinolinoline |
C9H8N2O |
45,78 |
160,173 |
20 |
21 |
|
1511784 |
7-chloro-5-nitroquinoline-8-ol |
C9H5ClN2O3 |
44,18 |
224,601 |
20 |
21 |
|
14125599 |
7-bromo-8-hydroxy-5-nitroquinoline |
C9H5BrN2O3 |
46,64 |
269,052 |
20 |
21 |
|
21823019 |
7-Fluor-5-nitroquinoline-8-ol |
C9H5FN2O3 |
34,02 |
208,146 |
20 |
21 |
|
44466195 |
5 Isopropylamino-quinolin-8-ol |
C12H14N2O |
78,48 |
202,252 |
29 |
30 |
|
45487202 |
5 Ethylamino-quinolin-8-ol |
C11H12N2O |
81,42 |
188,226 |
26 |
27 |
|
166150257 |
4-Difluoromethyl-7-nitroquinolin-8-ol |
C10H6F2N2O3 |
95,08 |
240,163 |
23 |
24 |
|
168070012 |
5-nitro-7-prop-2-enoxi-imino-metil-quinoline-8-ol |
C13H11N3O4 |
14,41 |
273,244 |
31 |
32 |
|
169355519 |
5-Isocianate-quinolin-6-ol |
C10H6N2O2 |
30,19 |
186,167 |
20 |
21 |
The use of the Proteins Plus web server allowed structural characterization of the cathepsin B binding pocket involved in catalysis. It predicts the key amino acids of the catalytic site and shows those involved in the catalytic activity of the enzyme. The results were taken into account for molecular docking studies. Twelve ligands were evaluated as possible inhibitors of cathepsin B using Autodock Tools. Table 2 shows the results obtained.
|
Table 2. Molecular docking results using Autodock Tools. The value of the ligand-receptor affinity energy is shown. Only the amino acids of the active site that were inhibited by each compound were taken into account. Results that are considered to be potential cathepsin B-inhibiting drugs are highlighted in red. ΔG represents the ligand-protein affinity, and Ki, the inhibition constant |
|||||
|
Linking |
ΔG (KJ/mol) |
Ki (µM) |
Inhibited key amino acids |
E. electrostatic (KJ/mol) |
E. torsional |
|
250068325 |
-5,64 |
- |
- |
0,33 |
0,89 |
|
199506 |
-7,27 |
4,69 |
His110, His111, Gly198 |
0,09 |
0,89 |
|
245295 |
-3,82 |
1,59 |
Gly198 |
1,05 |
0,89 |
|
416002 |
-4,79 |
307,43 |
His111, His199 |
0,04 |
0,60 |
|
1511784 |
-5,56 |
83,56 |
His111, His199, Gly198 |
0,24 |
0,60 |
|
14125599 |
-5,51 |
92,05 |
His111, His199, Gln23 |
0,37 |
0,60 |
|
21823019 |
-5,24 |
- |
- |
0,29 |
0,60 |
|
44466195 |
-5,39 |
- |
- |
0,15 |
0,89 |
|
45487202 |
-5,08 |
187,08 |
His111, His199, Gly198 |
0,04 |
0,89 |
|
166150257 |
-5,99 |
48,08 |
His199, Gln23, Gly198 |
0,27 |
0,89 |
|
168070012 |
-4,33 |
674,10 |
His111, His199 |
0,52 |
0,6 |
|
169355519 |
-5,26 |
140,27 |
His111, Gln23 |
0,09 |
0,6 |
From the data of 12 compounds, potential cathepsin B inhibitors were selected based on whether they inhibited key amino acids and had a reasonable inhibition constant. The compounds that met the above criteria were 1511784, 14125599, and 45487202. The hydrogen-bonding interactions between these ligands and the amino acids of the active site of cathepsin B are shown (Figure 2), as well as the distances between these bonds. Additionally, the energy value associated with each interaction is indicated.
From the use of Autodock Tools, the main interactions between the amino acids of the cathepsin B binding pocket and the ligands of interest were constructed (Figure 3). In each case, the ligand is visualized as a transparent solid sphere located in the center, and the amino acids as tubes. The interactions that are established are shown as spheres of cells.
Figure 2. Ligand-receptor interaction for compounds 1511784, 14125599, and 45487202. The energy values are shown in white color and green color, the binding distances between the atoms involved in the interactions. Lig: represents ligand; His: histidine; Gly, glycine and Gln, glutamine
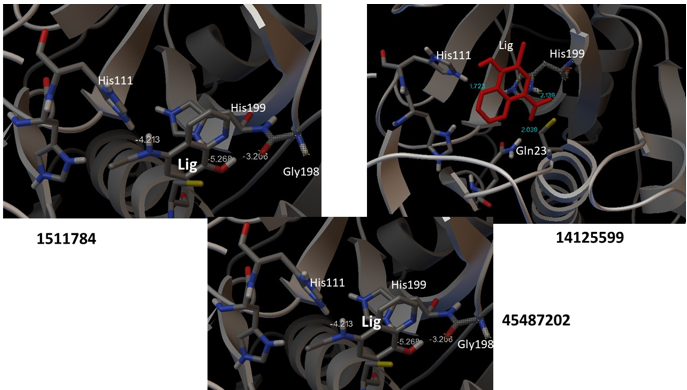
Figure 3. Interactions present between the ligands 1511784, 14125599, and 45487202 and the key amino acids for the catalysis process. The interactions are shown as spheres of cells. Each ligand is shown in the center
DISCUSSION
Cathepsin B is a lysosomal cysteine protease involved in tumor progression and metastasis. It represents a crucial target for the development of new antitumor agents. Inhibitors of proteases that are overexpressed in different carcinomas are currently gaining momentum. Especially the use of non-peptide compounds with inhibitory activity in silico and in vitro models has proven useful against tumor progression.(9) Nitroxoline has been found to have effective anticancer properties in breast, glioma, bladder, pancreatic, and prostate cancer.(10,11) It is involved in the activation of cell apoptosis, cell cycle arrest, and suppression of angiogenesis. Studies in animal models have shown that nitroxoline is an effective and reversible inhibitor of cathepsin B catalytic activity.(12,13)
Figure 1 shows the data of compounds extracted from the PubChem database. All are derived from the nitrosoline nucleus and were energy-minimized so that they were in a stable conformation analogous to their natural form. The GAFF force field was applied, compatible with possible drugs with C, H, O, N, F, and Cl, among other elements, in their structure. In this sense, Table 1 lists the energy values of each compound that was subjected to the GAFF field and its corresponding molecular formula. All the selected compounds had between 9 and 15 carbons.
Each ligand was evaluated in 10 different conformations at the cathepsin B site. However, the best value was taken in each case. The ligand-protein affinity is given by a scoring function involving the sum of the electrostatic and Van der Waals energies. The lower the affinity value, the more stable the compound. All values were below “0”, indicating that the ligand-receptor complex is stable and the interaction is more likely to occur. However, compounds 250068325, 21823019, and 44466195 were disregarded (Table 2), as they do not inhibit any amino acid residue of the catalytic site of cathepsin B. Ligands 245295, 416002, and 168070012 presented lower energy values with respect to the rest, indicating a lower stability of interaction with the active site of the receptor.
In this sense, the potential drugs selected were 1511784, 14125599, and 45487202, as both present binding affinities (ΔG=-5.56; -5.51 and -5.08, respectively) for cathepsin B and inhibit key amino acids of its binding pocket: Gln23, His 111, His199 and Gly 198. The results are similar to other in silico studies reported for cathepsin B in cancer and Alzheimer’s disease. In this one, the affinity energy for the inhibitors was -6.74 and -6.12 (KJ/mol). (14,15) His 199 is an essential amino acid in the catalysis of the catalytic triad of cysteine proteases, which Asn, Cys, and His form. Cys and His form an ionic pair in the catalytic dyad. His acts together with cysteine in the formation of the nucleophilic thiolate (thiol group of cysteine)-imidazole (His group) pair. Gln23 subsequently participates in the stabilization of the tetrahedral intermediate by the formation of a hydrogen bridge.(16)
Table 2 shows the inhibition constant (Ki) for the ligands that inhibited some amino acids from the catalytic site. The values were 83.56, 92.05, and 187.08 µM for potential inhibitors 1511784, 14125599, and 45487202. There are differences between the calculated and experimental Ki. However, this in silico value gives us a measure of the competitive, non-reversible inhibition that these compounds could have in vitro assays.
Figure 2 shows the hydrogen bonding interactions established between each ligand and cathepsin B, as well as the affinity energy of each interaction. In all cases, the ΔG value is negative, which evidences the binding stability of the complex formed. In the case of the ligand 14125599, the H-O bond distance is reported, where the values are between 1.6 to 2.0 Å. This indicates that the ligand behaves as a hydrogen acceptor, while the amino acid of the active site corresponds to the O or N donor.
Figure 3 shows the ligand-acceptor interactions. His 111 is responsible for dipeptidyl carboxypeptidase activity and acts together with His 110 in stabilizing the carboxyl terminus of the substrate peptide. His is located adjacent to small residues such as Gly or alanine (Ala). In this case, the adjacent position of His119 with respect to Gly198 is observed in Figure 2, ligand 1511784. The pharmacological candidates 1511784, 14125599, and 45487202 form hydrogen bonds with these amino acid residues, suggesting their inhibitory role in cathepsin B against breast cancer. The docking technique has some limitations as it considers the receptor (cathepsin B) as a rigid structure during the interaction. It presents a low sensitivity for large molecules and a maximum of 8 rotatable bonds.(17) In this case, small molecules having less than 5 rotatable bonds were analyzed.
The results of this computational study suggest that inhibitors 1511784, 14125599, and 45487202 exhibit binding affinities for the active site of cathepsin B with favorable interaction-free energy values. Inhibition of key residues for cathepsin B function is evidenced. This contributes to the search for new strategies to combat breast cancer, specifically in the detection of molecules derived from the nitrosoline nucleus. The simulations performed, despite presenting highly predictive results, underestimate the mode and time of action of the molecules, as well as the optimal concentrations of activity, which depends exclusively on experimental assays.
REFERENCES
1. Trayes KP, Cokenakes SE. Breast Cancer Treatment. American Family Physician. 2021 Aug; 104(2):171-8. Disponible en: https://www.aafp.org/pubs/afp/issues/2021/0800/p171.pdf
2. Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, et al. Breast Cancer Statistics. CA Cancer J Clin. 2022 Dec; 72(6):524–18. Disponible en: https://acsjournals.onlinelibrary.wiley.com/doi/pdf/10.3322/caac.2175
3. Waks A, Winer E. Breast cancer treatment a review. Rev Clinical Review and Education. 2019; 321(3): 3p. Disponible en: http://bdrc.tums.ac.ir/uploads/140/2020/Jun/17/Breast-Cancer-Treatment-Jan-2019-1.pdf
4. Ministerio de Salud Pública. Anuario estadístico 2019 La Habana. MINSAP. Disponible en: https://instituciones.sld.cu/ucmvc/files/2023/10/Anuario-Estad%C3%ADstico-de-Salud-2022-Ed-2023.pdf
5. Watkins EJ. Overview of breast cancer. Journal of the American Academy of Pas. 2019 Oct; 32(10):13-5. Disponible en: https://journals.lww.com/jaapa/fulltext/2019/10000/Overview_of_breast_cancer.3.aspx
6. De Dios DH. Selección de blanco terapéutico en la catepsina B para el desarrollo de fármacos contra el cáncer de mama. Rev Ciencias Médicas. 2021; 25(5): 10p. Disponible en: http://revcmpinar.sld.cu/index.php/publicaciones/article/view/5003
7. Aggarwal N, Sloane B. Cathepsin B: Multiple roles in cancer. Rev Proteomics Clin. Appl. 2014; 8(5): 427-11. Disponible en: https://onlinelibrary.wiley.com/doi/abs/10.1002/prca.201300105 7.
8. Gondi C, Rao J. Cathepsin B as a cancer target. Rev Expert Opin Ther Targets. 2013; 17(3): 281–11. Disponible en: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3587140/
9. Sosic I, Mitrovic´ A, Hrvoje C, Knez D, Brodnik HZ, Štefane B et al. Cathepsin B inhibitors: Further exploration of the nitroxoline core. Bioorganic & Medicinal Chemistry Letters. 2018; 28: 1239–9. Disponible en: https://www.sciencedirect.com/science/article/abs/pii/S0960894X18301471
10. Xu N, Huang L, Li X, Watanabe M, Li C, Xu A et al. The Novel Combination of Nitroxoline and PD-1 Blockade, Exerts a Potent Antitumor Effect in a Mouse Model of Prostate Cancer. International Journal of Biological Sciences. 2019; 15(5): 919-9. Disponible en: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6535792/
11. Xu N, Lin W, Sun J, Sadahira T, Xu A, Watanabe M et al. Nitroxoline inhibits bladder cancer progression by reversing EMT process and enhancing anti-tumor immunity. Journal of Cancer. 2020; 11(22): 6633-6641.
12. Mirkovic B, Markelc B, Butinar M, Mitrovic A, Sosic I, Gobec S, et al. Nitroxoline impairs tumor progression in vitro and in vivo by regulating cathepsin B activity. Oncotarget. 2015; 6: 19027-42. Disponible en: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4662473/
13. Mitrovic A, Kljun J, Sosic I, Ursic M, Meden A, Gobec S et al. Organoruthenated Nitroxoline Derivatives Impair Tumor Cell Invasion through Inhibition of Cathepsin B Activity. Inorganic Chemistry. 2019; 58(18): 12334-12347. Disponible en: https://pubs.acs.org/doi/full/10.1021/acs.inorgchem.9b01882
14. Ranjbar D, Reza M, Dehghani Z, Firuzi D, Edraki N, Khoshneviszadeh M. Dihydronaphthalenone chalconoid derivatives as potential cathepsin B inhibitors; design, synthesis, cytotoxicity evaluation and docking analysis. Braz. J. Pharm. Sci. 2021;57: 1-14. Disponible en: https://www.scielo.br/j/bjps/a/GTb8RkyrDwSRSbKCpRBnSMK/?lang=en
15. Chitranshi N, Kumar A, Sheriff S, Gupta V, Godinez A, Saks D et al. Identification of Novel Cathepsin B Inhibitors with Implications in Alzheimer’s Disease: Computational Refining and Biochemical Evaluation. Cells. 2021; 10(1946). Disponible en: https://www.mdpi.com/2073-4409/10/8/1946
16. Naranjo F. Análisis in silico de la catepsina B de Fasciola hepática como diana terapéutica. REDVET [Internet]. 2009 [citado: 08/03/2021]; 10(2): 1-36. Disponible en: https://www.redalyc.org/pdf/636/63617114011.pdf
17. Guadalupe W, Paucara B, Grados, R. Acomplamiento molecular: criterios prácticos para la selección de ligandos biológicamente activos e identificación de nuevos blancos terapéuticos. Rev.Cs.Farm. y Bioq. 2019; 7(2). Disponible en: http://www.scielo.org.bo/scielo.php?pid=S2310-02652019000200006&script=sci_arttext
CONFLICT OF INTEREST
The author declares that there is no conflict of interest.
AUTHOR CONTRIBUTION
The author participated in the conceptualization, drafting - initial draft, drafting - revision and editing.
FUNDING
The author did not receive funding for the development of this editorial.